Contents
- Properties of K-Space
- Center of K-Space
- K-space lines
- Center projections
- The Image is the (inverse) 2DFT of the Raw data
- Same idea with Central 128 lines
- Images of 512 resolution are rare. We will mostly work with 256x256
- remove unused stuff
- The high k values (periphery of k-space) contain high SPATIAL frequency information.
- We can smooth less aggressively by making attenuating the high k values less.
- Construct high pass filter
- Common artifacts
- Two or more spikes creates a sort of herringbone pattern
- Saturation
- Apodization.
- More aggressive apodization happens in long readout scans such as EPI
- Fourier Shift theorem
- Motion artifact simulation
Properties of K-Space
These exercise are intended to give you a sense of the properties of K-space (the MR raw data) and the ways in which the artifacts and behavior of MR images depends on the k-space trajectories and dadta
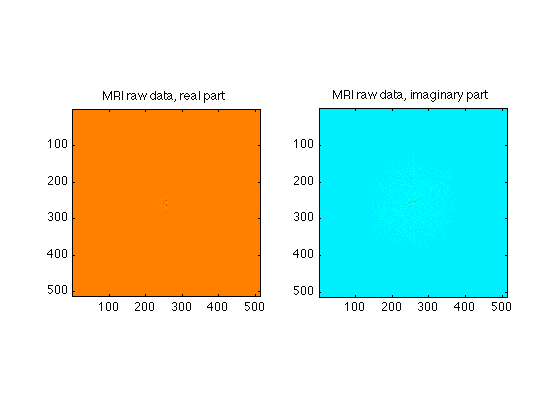
clear; % clears data from memory clf; % clears the figures load RawData; % This is a file containing raw data for a 512 X 512 MR image whos; % shows you the characteristics of the input data mygray = [0:1/255:1;0:1/255:1;0:1/255:1]'; % Supplements MATLAB's default 64 level gray scale [ydim, xdim] = size(RawData); % First, let's look at the MR raw data, which has both real and imaginary % parts. subplot(1,2,1); image(real(RawData),'CdataMapping','scaled'); axis square; set(gcf,'Color','White'); colormap('jet'); title('MRI raw data, real part'); subplot(1,2,2); image(imag(RawData),'CdataMapping','scaled'); axis square; title('MRI raw data, imaginary part'); % Note that very little of the signal appears outside of the center regions % of k=space
Name Size Bytes Class Attributes RawData 512x512 4194304 double complex
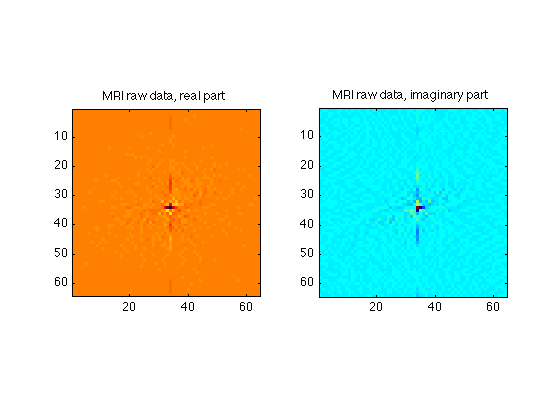
Center of K-Space
We can zoom in on the center 64 X 64 points of k-space to see the data a bit better
Ymid = ydim/2; Xmid = xdim/2; Center64 = RawData( Ymid-32:Ymid+31, Xmid-32:Xmid+31 ); subplot(1,2,1); image(real(Center64),'CdataMapping','scaled'); axis square; set(gcf,'Color','White'); colormap('jet'); title('MRI raw data, real part'); subplot(1,2,2); image(imag(Center64),'CdataMapping','scaled'); axis square; title('MRI raw data, imaginary part');
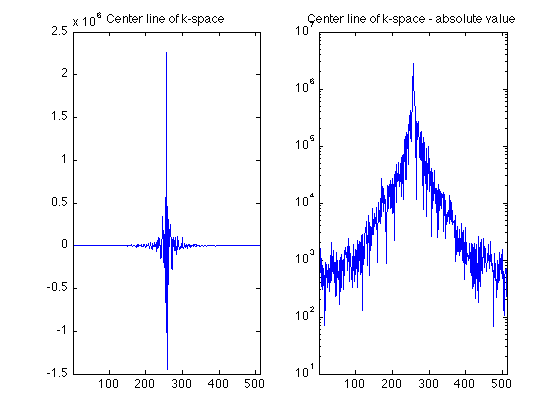
K-space lines
As individual lines, you will see that most look a bit like sinc functions.
clf; subplot(1,2,1); plot(real(RawData(256,:))); set(gca,'xlim',[1 512]); title('Center line of k-space'); subplot(1,2,2); plot(abs(RawData(256,:))); set(gca,'xlim',[1 512]); set(gca, 'yscale','log'); title('Center line of k-space - absolute value');
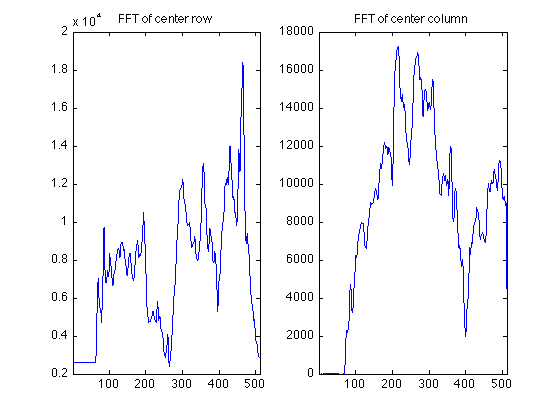
Center projections
The 1D Fourier Transform of the center LINE of K-space is euivalent to the project shadow of the object in the imager along the x axis.
clf; subplot(1,2,1); centerProjectionRow = ifft(fftshift(RawData(256,:))); % The raw data for a REAL object in the scanner generally is complex, % containing both real and imaginary parts. However, the mechanics of the % instrument, and effects such as subject motion, actually produce raw data % result in raw data whose fourier transform is complex. Thus we generally % look at the magnitude (absolute value) of the images. plot(abs(centerProjectionRow)); set(gca,'xlim',[1 512]); title('FFT of center row'); % The 1D Fourier Transform of the center COLUMN of K-space is equivalent to % the project shadow of the object in the imager along the Y axis. subplot(1,2,2); centerProjectionCol = ifft(fftshift(RawData(:,256))); plot(abs(centerProjectionCol)); set(gca,'xlim',[1 512]); title('FFT of center column');
The Image is the (inverse) 2DFT of the Raw data
clf; ReconImage = ifftn(fftshift(RawData)); subplot(1,2,1); % Note that we generally use the absolute value of the raw data to % accomodate phase variations from factors such as shim homogeneity. image(abs(ReconImage), 'CdataMapping','direct'); % I use the direct lookup table so that we can compare intensities axis image; colormap(mygray); % A lower resolution image is formed from the central area of k-space Center64Image = ifftn(fftshift(Center64)); subplot(1,2,2); image(abs(Center64Image/64), 'CdataMapping','direct'); % The scaling has to do with integrating a smaller area of k-space axis image; title('64X64 matrix');

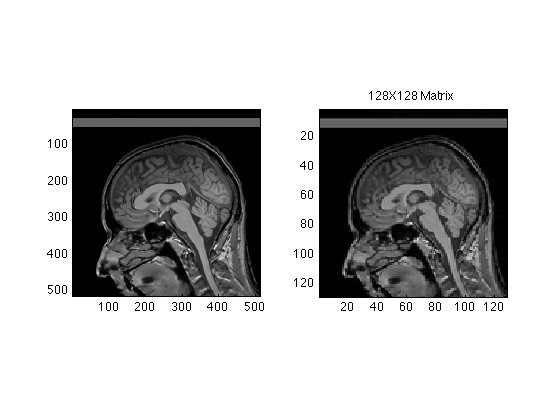
Same idea with Central 128 lines
Center128 = RawData( Ymid-64:Ymid+63, Xmid-64:Xmid+63 ); Center128Image = ifftn(fftshift(Center128)); subplot(1,2,2); image(abs(Center128Image/16), 'CdataMapping','direct'); axis image; title('128X128 Matrix');
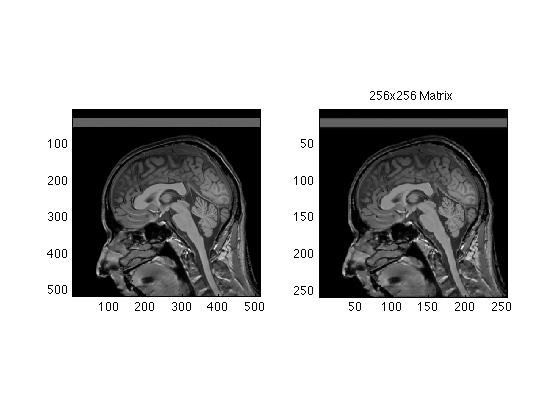
Images of 512 resolution are rare. We will mostly work with 256x256
Raw256 = RawData( 129:384, 129:384 ); % for later Center256Image = ifftn(fftshift(Raw256)); subplot(1,2,2); image(abs(Center256Image/4), 'CdataMapping','direct'); axis image; title('256x256 Matrix');
remove unused stuff
clear Center64 Center64Image Center128 Center128Image
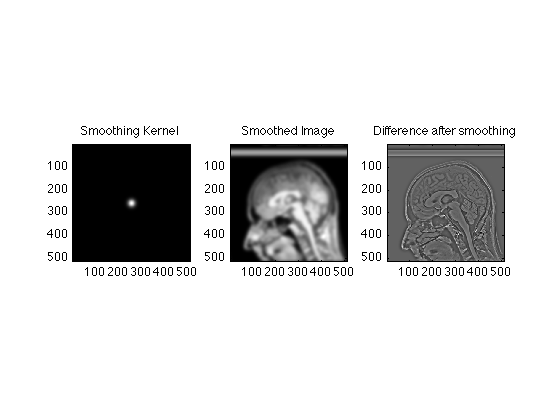
The high k values (periphery of k-space) contain high SPATIAL frequency information.
Attenuating these makes tends to smooth the image. Here, we smoothly attenuate these values by multiplying their intensities by a two dimensional gaussian.
% Create the gaussian: x=0:511; SmoothingWidth = 32; SigmaSquared = (xdim/SmoothingWidth)^2; g=repmat(exp(-(x-256).^2/SigmaSquared),512,1); TwoDGauss = g' .* g; clf; subplot(1,3,1); image(abs(TwoDGauss),'CDataMapping','scaled'); title('Smoothing Kernel'); axis image; % Multiply the k data: FSmoothRaw = TwoDGauss .* RawData; % Create a new image SmoothedRecon = ifftn(fftshift(FSmoothRaw)); subplot(1,3,2); image(abs(SmoothedRecon), 'CdataMapping','scaled'); title('Smoothed Image'); axis image; % Plot the difference between the smoothed and original image subplot(1,3,3); image(abs(ReconImage) - abs(SmoothedRecon), 'CdataMapping','scaled'); title('Difference after smoothing'); axis image; % Notice that the difference image is mostly edges
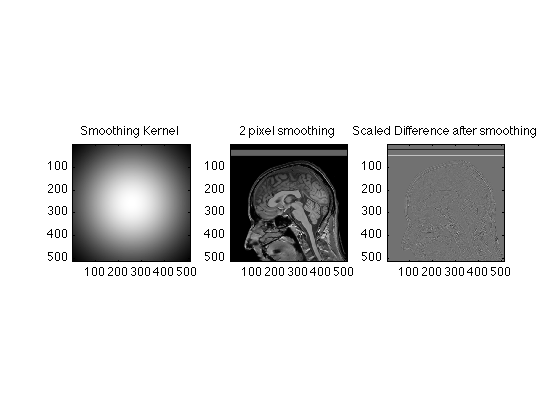
We can smooth less aggressively by making attenuating the high k values less.
SmoothingWidth = 2; SigmaSquared = (xdim/SmoothingWidth)^2; g=repmat(exp(-(x-256).^2/SigmaSquared),512,1); TwoDGauss = g' .* g; subplot(1,3,1); image(abs(TwoDGauss),'CDataMapping','scaled'); title('Smoothing Kernel'); axis image; FSmoothRaw = TwoDGauss .* RawData; SmoothedRecon = ifftn(fftshift(FSmoothRaw)); subplot(1,3,2); image(abs(SmoothedRecon), 'CdataMapping','scaled'); title('2 pixel smoothing'); axis image; subplot(1,3,3); image(abs(ReconImage) - abs(SmoothedRecon), 'CdataMapping','scaled'); title('Scaled Difference after smoothing'); axis image; % Here, only the finest details were lost. % All of these exercises are actually examples of the Fourier convolution % theorem, by the way. Ask if you want to know more.
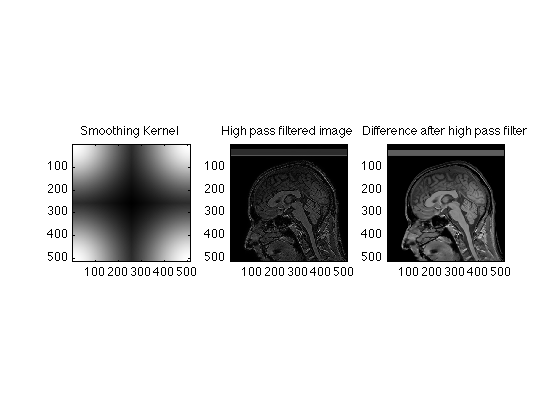
Construct high pass filter
It is just as easy to construct a high pass (edge) filter. Instead of emphasizing the low spatial frequencies we supress them, and use the high spatial frequencies instead.
% We start with a gaussian SmoothingWidth = 2.5; SigmaSquared = (xdim/SmoothingWidth)^2; g=repmat(exp(-(x-256).^2/SigmaSquared),512,1); TwoDGauss = g' .* g; % We then use the fftshift command reorder the data HighPass = fftshift(TwoDGauss); subplot(1,3,1); image(abs(HighPass),'CDataMapping','scaled'); title('Smoothing Kernel'); axis image; FHPRaw = HighPass .* RawData; HighPassRecon = ifftn(fftshift(FHPRaw)); subplot(1,3,2); image(abs(HighPassRecon), 'CdataMapping','scaled'); title('High pass filtered image'); axis image; subplot(1,3,3); image(abs(ReconImage) - abs(HighPassRecon), 'CdataMapping','direct'); title('Difference after high pass filter'); axis image; % Exercises % % Calculate and show the differences in images acquired with matrix sizes % of 64X64, 128X128, 256X256 and the original 512X512 image. % % Compare the effects of aggressive smoothing on the 512X512 image and the % 64X64 image. % % Explain the presence of a dark line in the spinal cord on the 128X128 % image. % % Construct an edge enhanced image
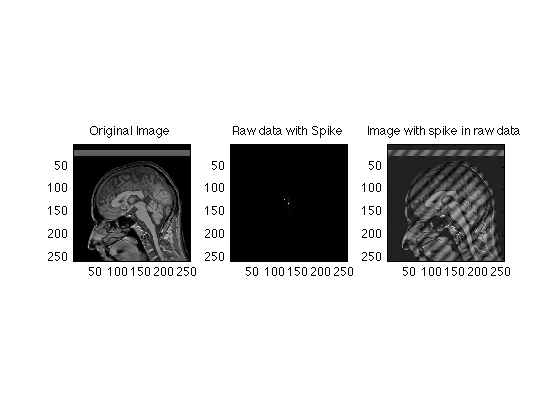
Common artifacts
It is not unusual for a single raw data point to become corrupted from events such as a small static electricity discharge. Here we simulation corruption of a single raw data time point
clear; mygray = [0:1/255:1;0:1/255:1;0:1/255:1]'; % Supplements MATLAB's default 64 level gray scale load RawData; Raw256 = RawData(129:384, 129:384); clear RawData; Spike256 = Raw256; Spike256(120,120) = 1e7; clf; subplot(1,3,1); imagesc(abs(ifftn(fftshift(Raw256)))); colormap(mygray); title('Original Image'); axis image; subplot(1,3,2); imagesc(abs(Spike256)); title('Raw data with Spike'); axis image; subplot(1,3,3); imagesc(abs(ifftn(fftshift(Spike256)))); title('Image with spike in raw data'); axis image; % Because the images are rectified (absolute value) spikes contaminate the % whole image
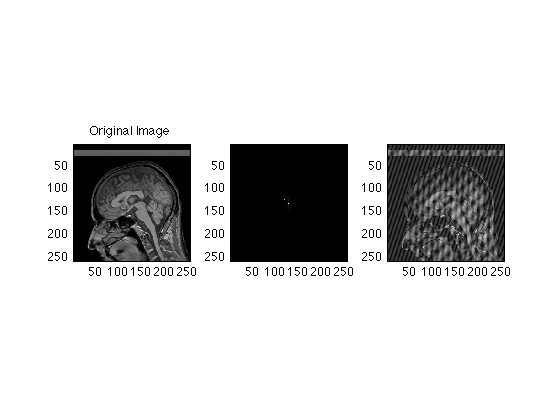
Two or more spikes creates a sort of herringbone pattern
Spike256(130,150) = 1e7; subplot(1,3,2); imagesc((abs(Spike256))); axis image; subplot(1,3,3); imagesc(abs(ifftn(Spike256))); axis image;
Saturation
clear Spike256; % Like all electronic devices, the MRI scanner has certain hardware % limitations. If, for example, the input levels are too high, the input % amplifiers will "clip". This can and does happen when the pre-scan is not % completed properly. % We can simulate this easily by setting a maximum signal intensity % threshold lower than the actual signal amplitude clf; subplot(2,2,1); imagesc(abs(ifftn(fftshift(Raw256)))); axis image; Clip = reshape( Raw256, 256*256,1 ); % convert to a linear vector for convenience; MaxReal = max(abs(real(Clip))); % Find the maxima separately in the real and imaginary parts MaxImag = max(abs(imag(Clip))); DataMax = max( MaxReal, MaxImag); Thresh = 0.2 * DataMax; % Set the threshold to 50% of the actual value count = 0; R=0; I=0; for pt = 1:size(Clip) rpt = real(Clip(pt)); ipt = imag(Clip(pt)); if( abs(rpt)> Thresh || abs(ipt)>Thresh) if abs(rpt) > Thresh R = abs(Thresh/rpt) * rpt; count = count+1; end if abs(ipt) > Thresh I = abs(Thresh/ipt) * ipt; count = count+1; end Clip(pt) = R + 1i*I; end end Clip = reshape(Clip, 256, 256); % Back to square raw data subplot(2,2,2); imagesc(abs(ifftn(Clip))); axis image; subplot(2,2,3); stairs(1:256,real(Raw256(130,:))); hold all; stairs(1:256,real(Clip(130,:))); subplot(2,2,4); stairs(1:256,imag(Raw256(130,:))); hold all; stairs(1:256,imag(Clip(130,:)));
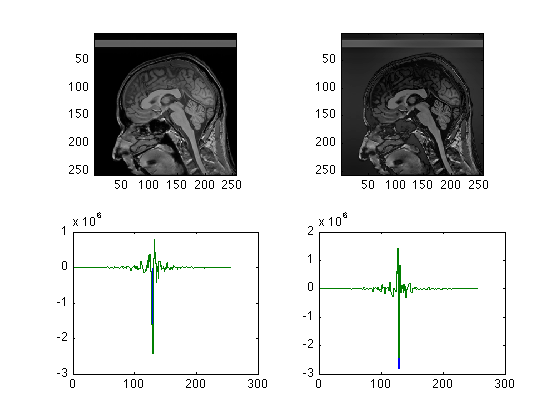
Apodization.
This section simulates, crudely, the effect of T2 star signal decay during readout.
clear; clf; load RawData; mygray = [0:1/255:1;0:1/255:1;0:1/255:1]'; % Supplements MATLAB's default 64 level gray scale % we will work here with the more conventional 256 X 256 images [ydim, xdim] = size(RawData); Ymid = ydim/2; Xmid = ydim/2; RawData = RawData(129:384, 129:384)/4; % again, the factor fo 4 intensity scaling comes from the smaller than original matrix size. [ydim, xdim] = size(RawData); Ymid = ydim/2; Xmid = ydim/2; ReconImage = ifftn(fftshift(RawData)); ReadoutDuration = 16; % msec T2star = 50; % msec Samples = (0:xdim-1) * ReadoutDuration / (T2star * (xdim-1)) ; Apodizer = exp(-Samples); Time = 0:ReadoutDuration/(xdim-1):ReadoutDuration; clf; plot(Time,Apodizer); set(gca,'ylim',[0 1]); title('T2 decay during readout'); pause(5) % Apply the weighting to each line of the original data. ApodizedData = repmat(Apodizer,ydim,1) .* RawData; ApodizedImage = ifftn(fftshift(ApodizedData)); subplot(1,3,1); image( abs(ReconImage), 'CdataMapping','direct' ); axis image; title('Original Image'); subplot(1,3,2); image(abs(ApodizedImage), 'CdataMapping','direct'); colormap(mygray); axis image; title('Apodized image'); subplot(1,3,3); image( abs(ReconImage - abs(ApodizedImage)), 'CdataMapping','direct' ); title('Difference after apodization'); axis image;
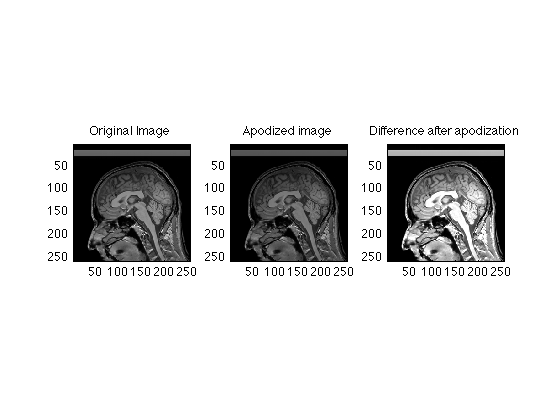
More aggressive apodization happens in long readout scans such as EPI
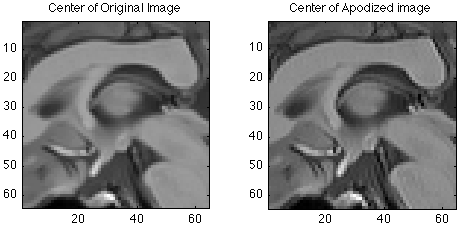
ReadoutDuration = 128; % msec T2star = 32; % msec Samples = (0:xdim-1) * ReadoutDuration / (T2star * (xdim-1)) ; Apodizer = exp(-Samples); Time = 0:ReadoutDuration/(xdim-1):ReadoutDuration; clf; plot(Time,Apodizer); set(gca,'ylim',[0 1]); title('T2 decay during readout'); pause(5); ApodizedData = repmat(Apodizer,ydim,1) .* RawData; ApodizedImage = ifftn(fftshift(ApodizedData)); subplot(1,3,1); image(abs(ReconImage), 'CdataMapping','scaled'); colormap(mygray); title('Original Image'); axis image; subplot(1,3,2); image( abs(ApodizedImage), 'CdataMapping','scaled' ); title('Apodized image'); axis image; subplot(1,3,3); image( abs(ReconImage - abs(ApodizedImage)), 'CdataMapping','direct' ); title('Difference after apodization'); axis image; % Here is a zoomed view pause(5) clf; subplot(1,2,1); imagesc(abs(ReconImage(Ymid-32:Ymid+31, Xmid-32:Ymid+31))); title('Center of Original Image'); axis image; subplot(1,2,2); imagesc(abs(ApodizedImage(Ymid-32:Ymid+31, Xmid-32:Ymid+31))); title('Center of Apodized image'); axis image;
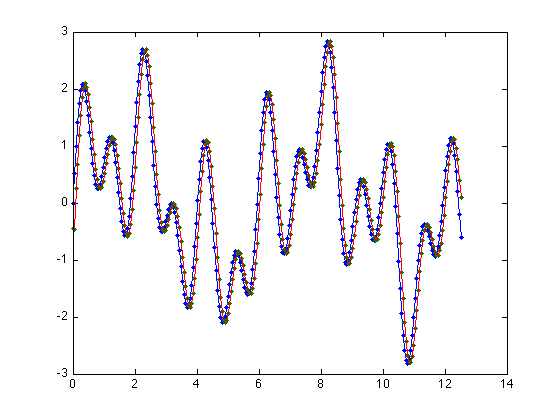
Fourier Shift theorem
t = 0:pi/64:4*pi-pi/64; s1 = sin(t); s3 = sin(3.3*t); s5 = sin(2*pi*t); s = s1+s3+s5; Fs = fft(s); a = .002*pi; S = -127:128; m = exp(-2*pi*1i*S*a); shifted = m .* fftshift(Fs); shiftedSignal = ifft(fftshift(shifted)); clf; pause(1); plot(t,(s),'.'); hold; plot(t,(s)); hold all; plot(t,(shiftedSignal),'.'); plot(t,(shiftedSignal));
Current plot held Warning: Imaginary parts of complex X and/or Y arguments ignored Warning: Imaginary parts of complex X and/or Y arguments ignored
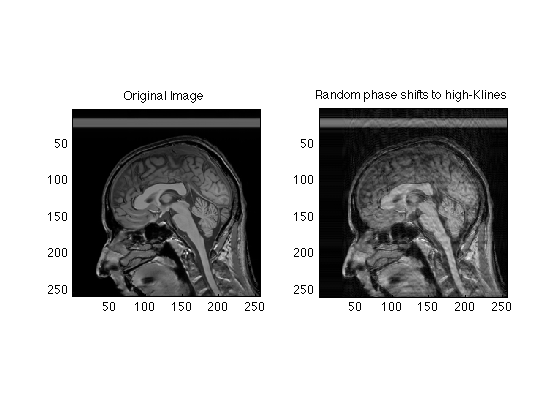
Motion artifact simulation
clf; clear; % The following initializes the randomization each time RandStream.setDefaultStream(RandStream('mt19937ar','seed',sum(100*clock))); mygray = [0:1/255:1;0:1/255:1;0:1/255:1]'; load RawData; Raw256 = RawData(128:384, 129:384);% work with a 256 x 256 image subplot(1,2,1); imagesc(abs(ifftn(fftshift(Raw256)))); axis square; colormap(mygray); % a single line for convenience in cut and paste title('Original Image'); [ys xs] = size(Raw256); C = Raw256; % we will corrupt the phases slightly for line = 1:ys; shift = exp(1i*0.5*randn()); % adds a random phase shift to the lines C(line,:) = Raw256(line,:) * shift; end subplot(1,2,2); imagesc(abs(ifftn(fftshift(C)))); title('Random phase shift'); axis square; colormap(mygray); disp('Click on Image'); waitforbuttonpress; C = Raw256; % we will corrupt a few lines near the center of k-space for line = 120:138; shift = exp(1i*0.5*randn()); % adds a random phase shift to the middle lines C(line,:) = Raw256(line,:) * shift; end subplot(1,2,2); imagesc(abs(ifftn(fftshift(C)))); title('Random phase shifts to middle lines'); axis square; colormap(mygray); disp('Click on Image'); waitforbuttonpress; C = Raw256; % we will corrupt a few lines for line = [64:90, 146:168] shift = exp(1i*3*randn()); % adds a random phase shift to the middle lines C(line,:) = Raw256(line,:) * shift; end subplot(1,2,2); imagesc(abs(ifftn(fftshift(C)))); title('Random phase shifts to high-Klines'); axis square; colormap(mygray);
Click on Image Click on Image